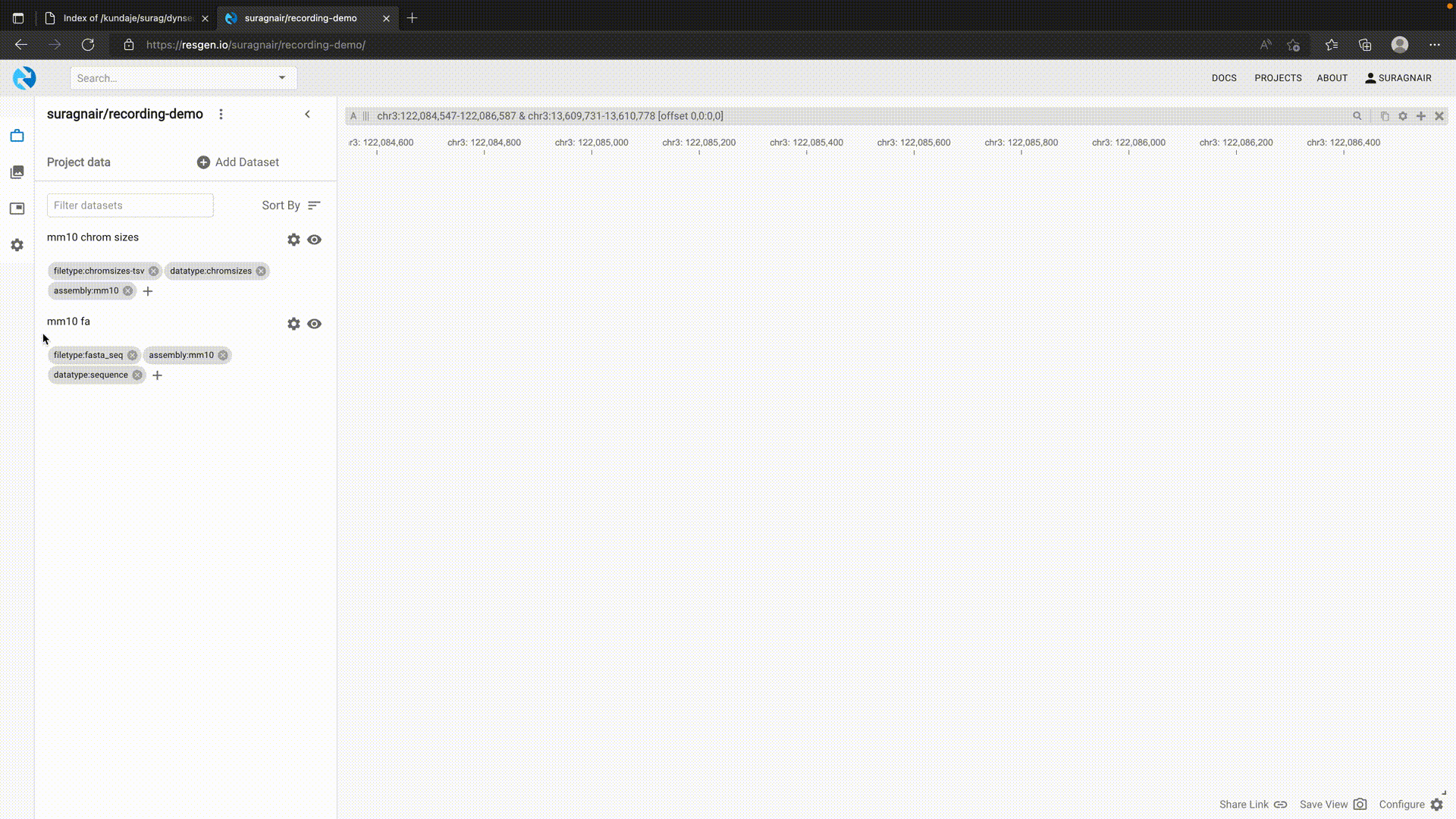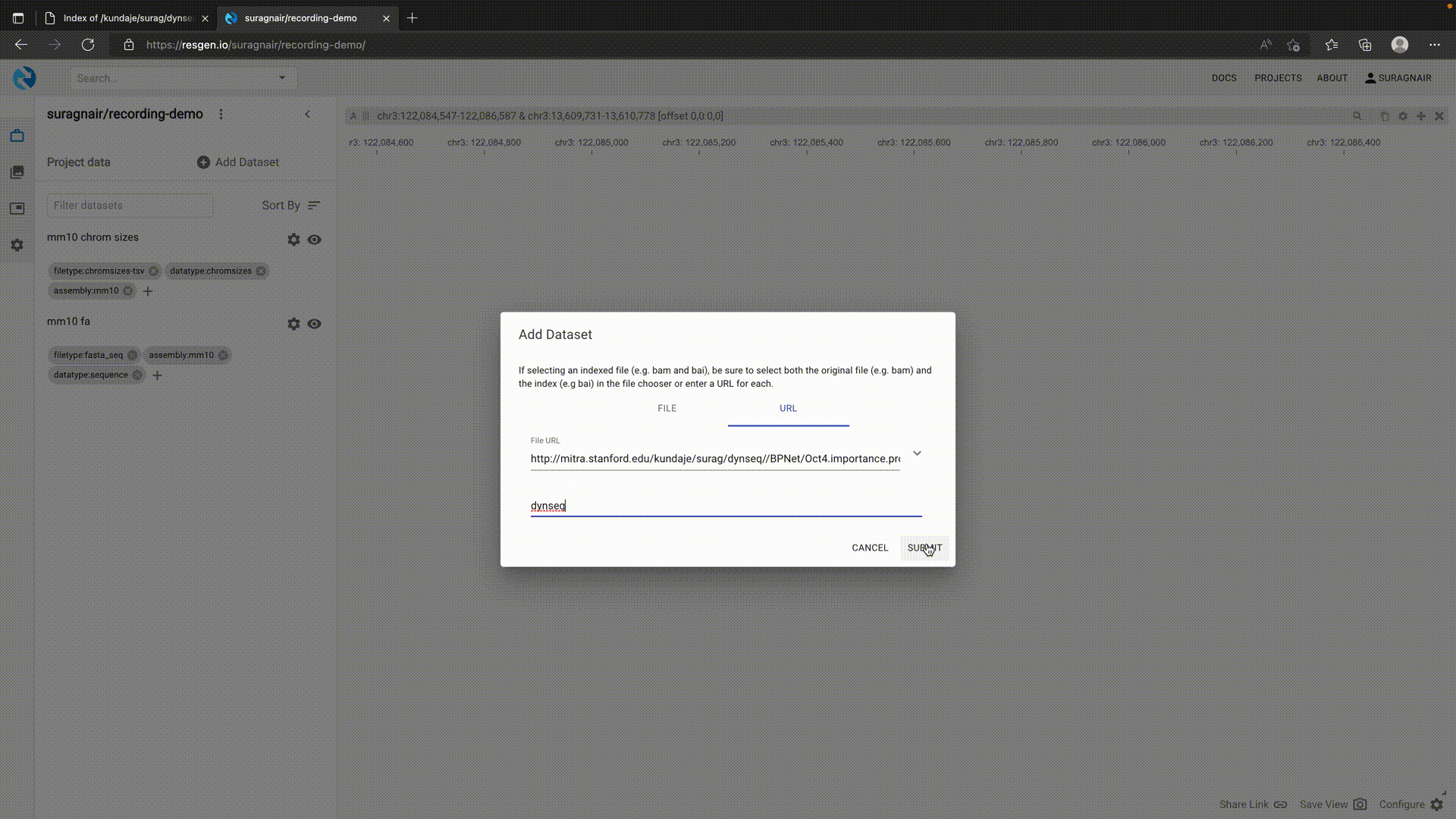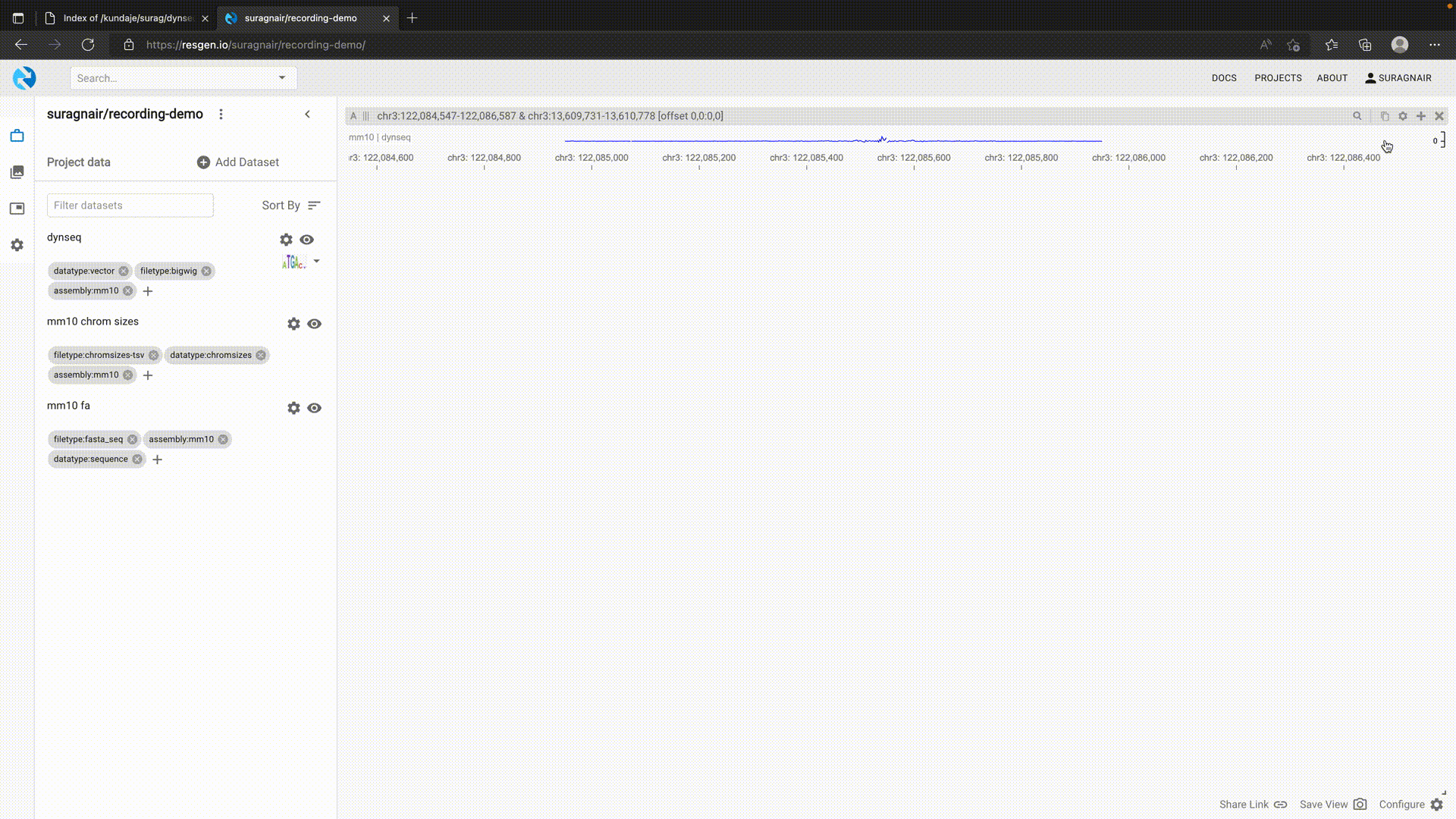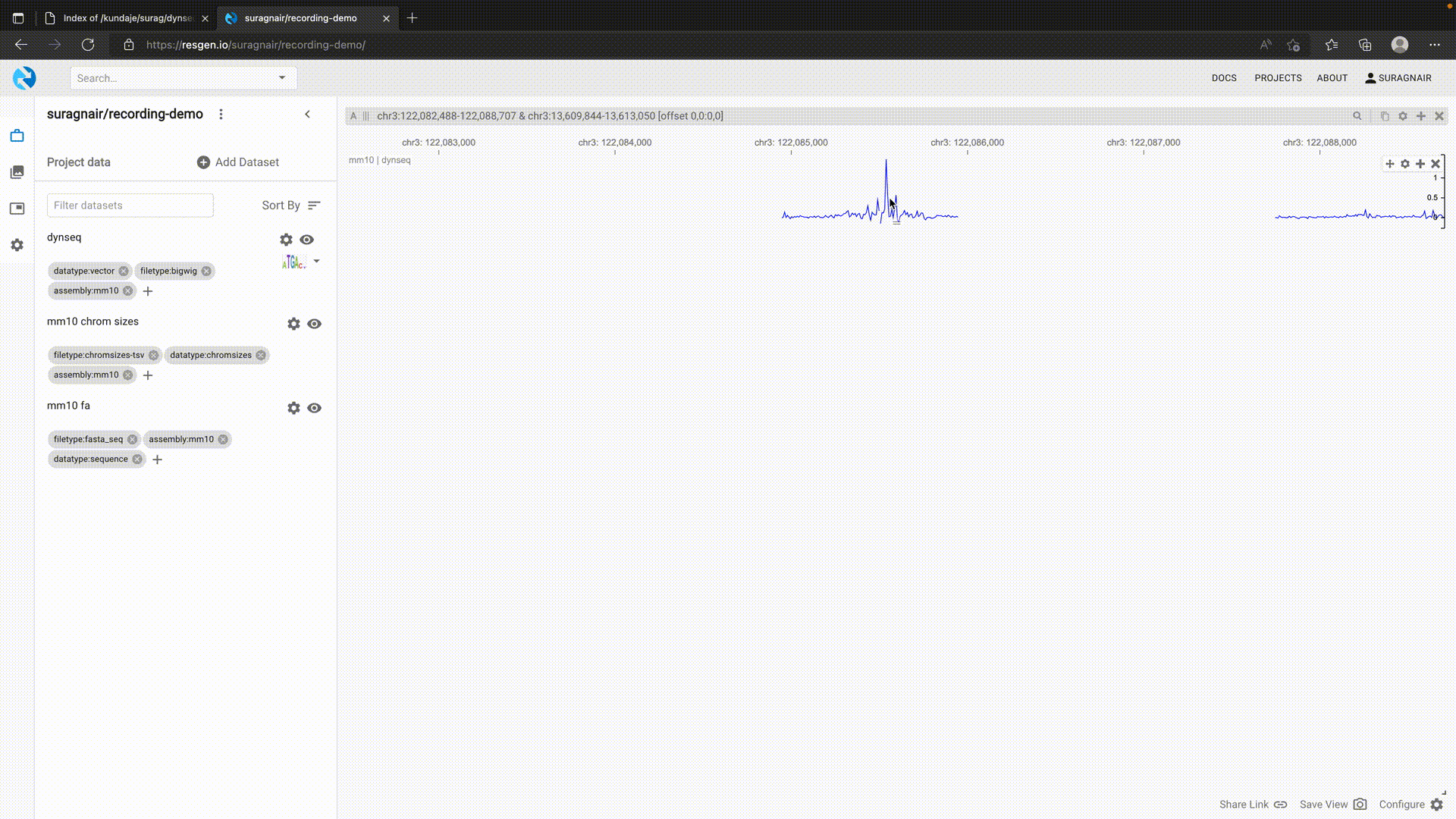
dynseq tracks
The dynseq genome browser track displays nucleotide characters scaled by user-specified, base-resolution scores provided in the BigWig format. It is currently supported by the WashU Epigenome Browser, UCSC Genome Browser, and HiGlass/Resgen. Instructions and details are provided below.
WashU Epigenome Browser
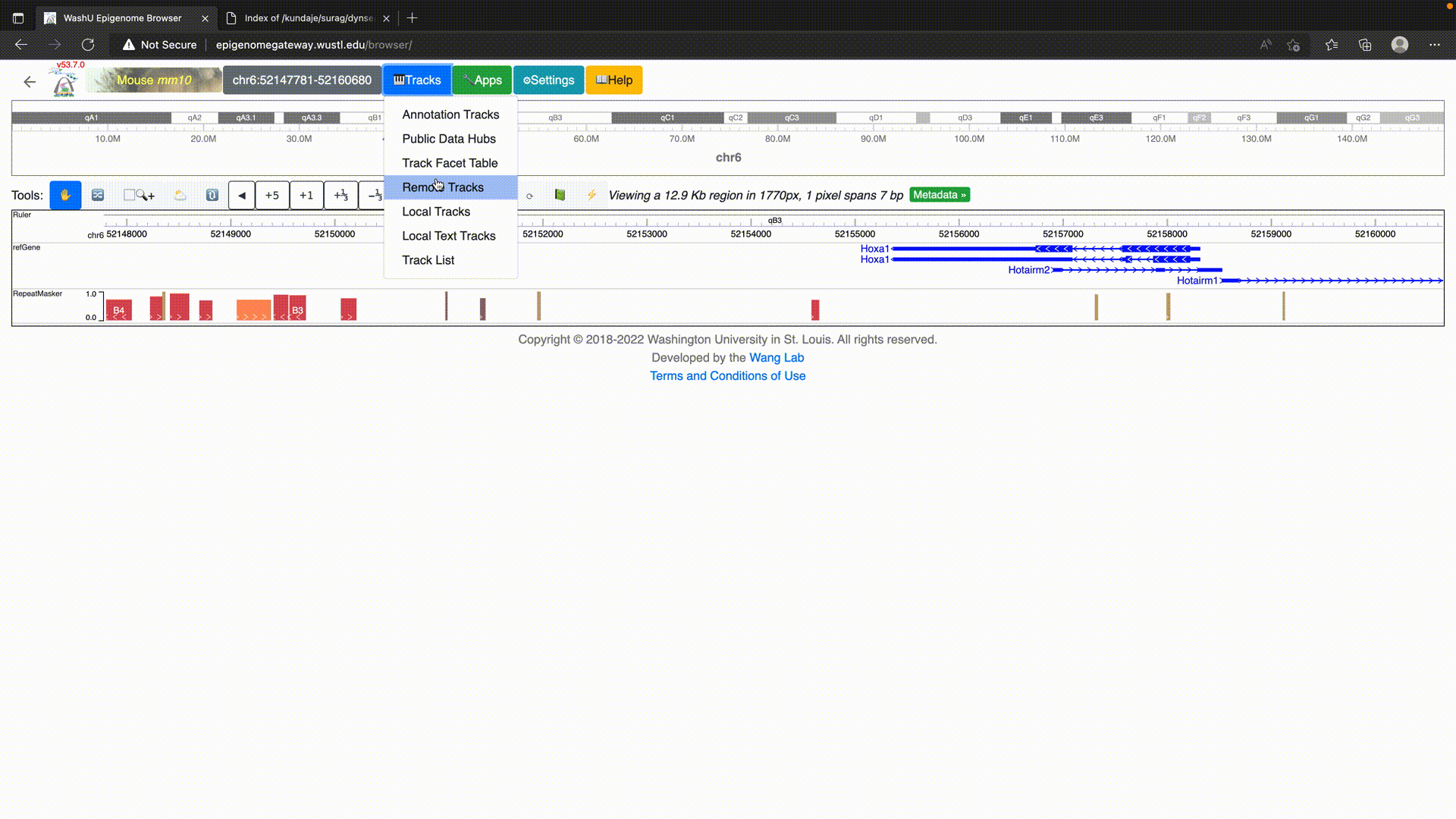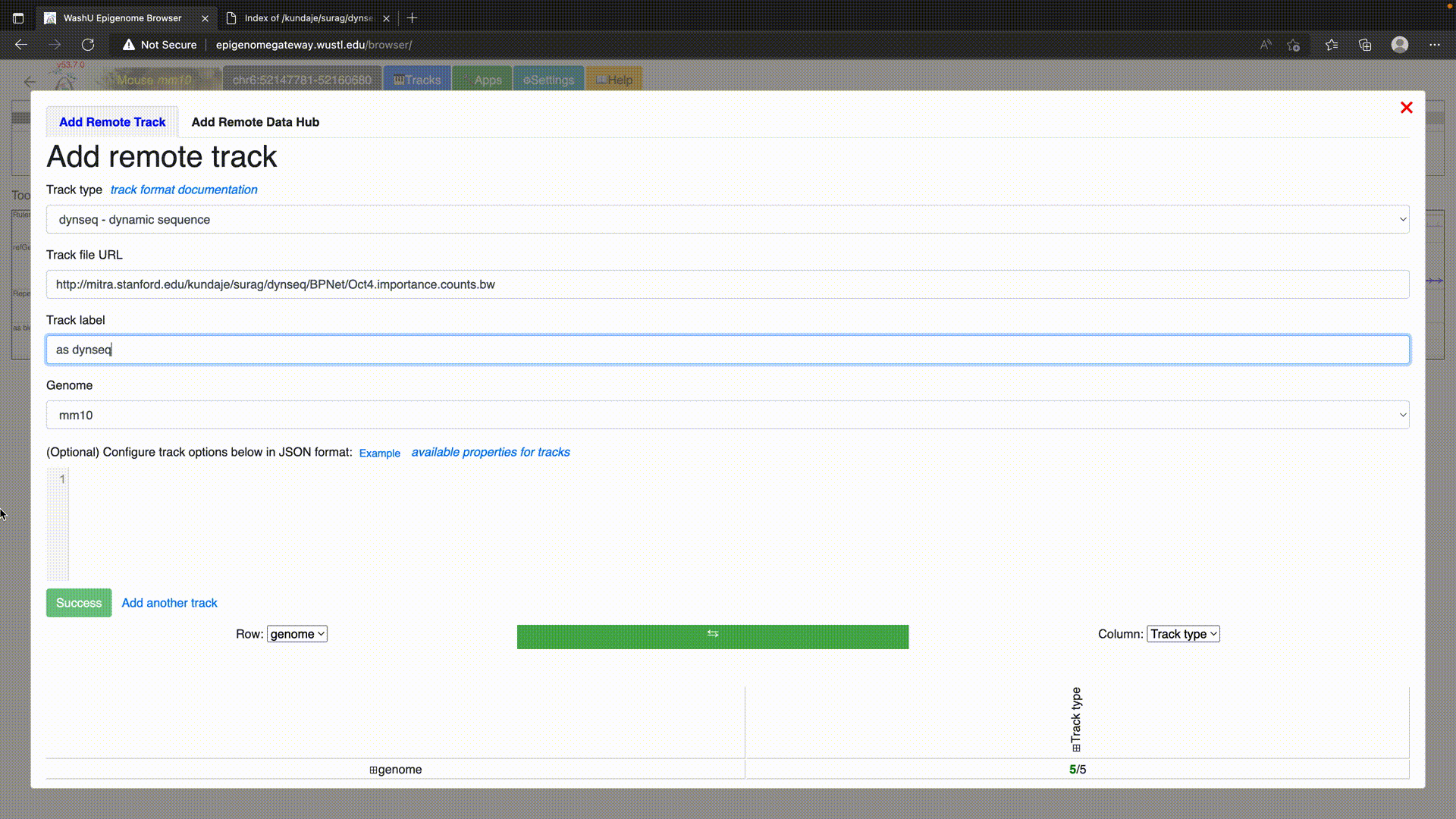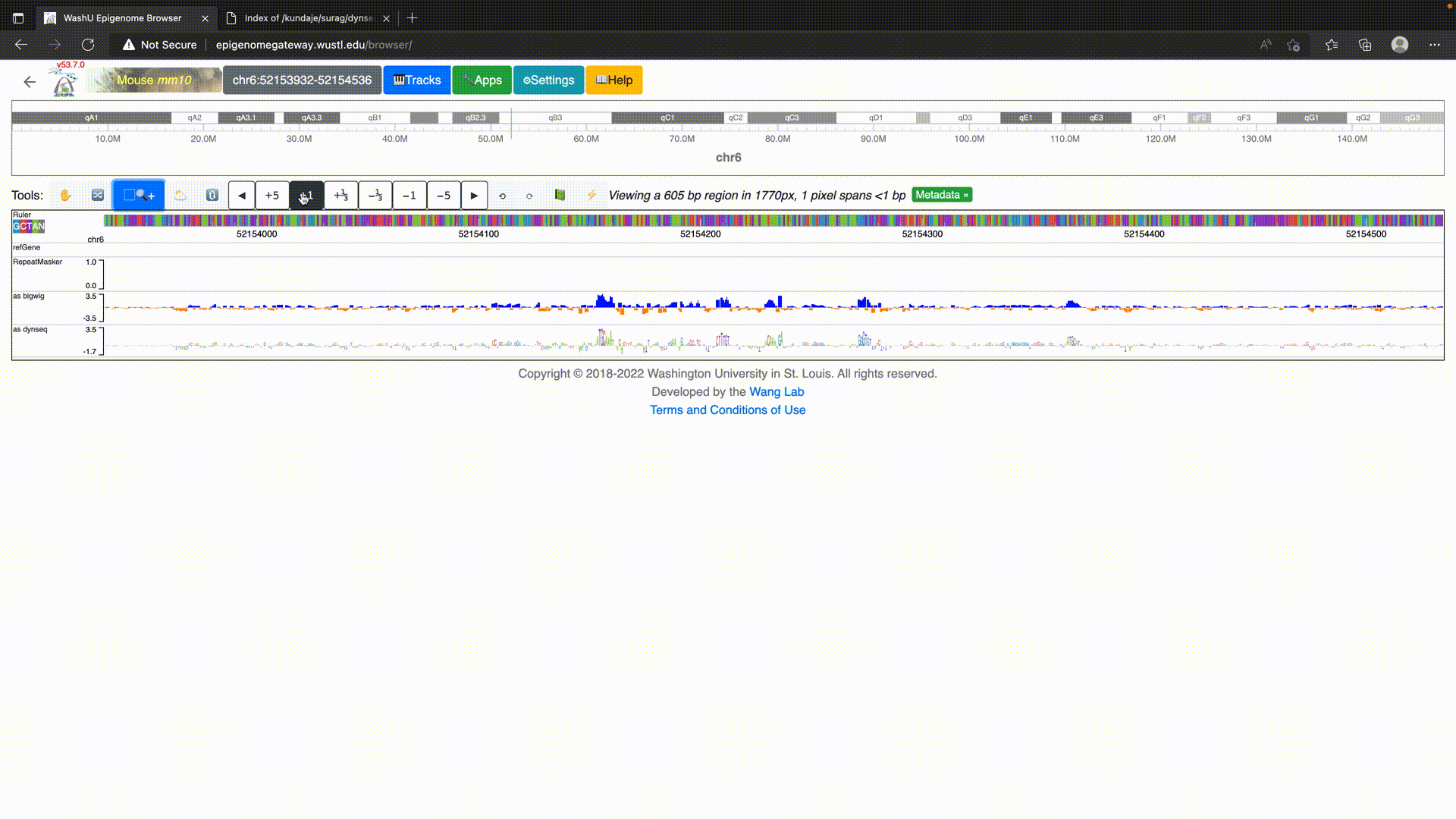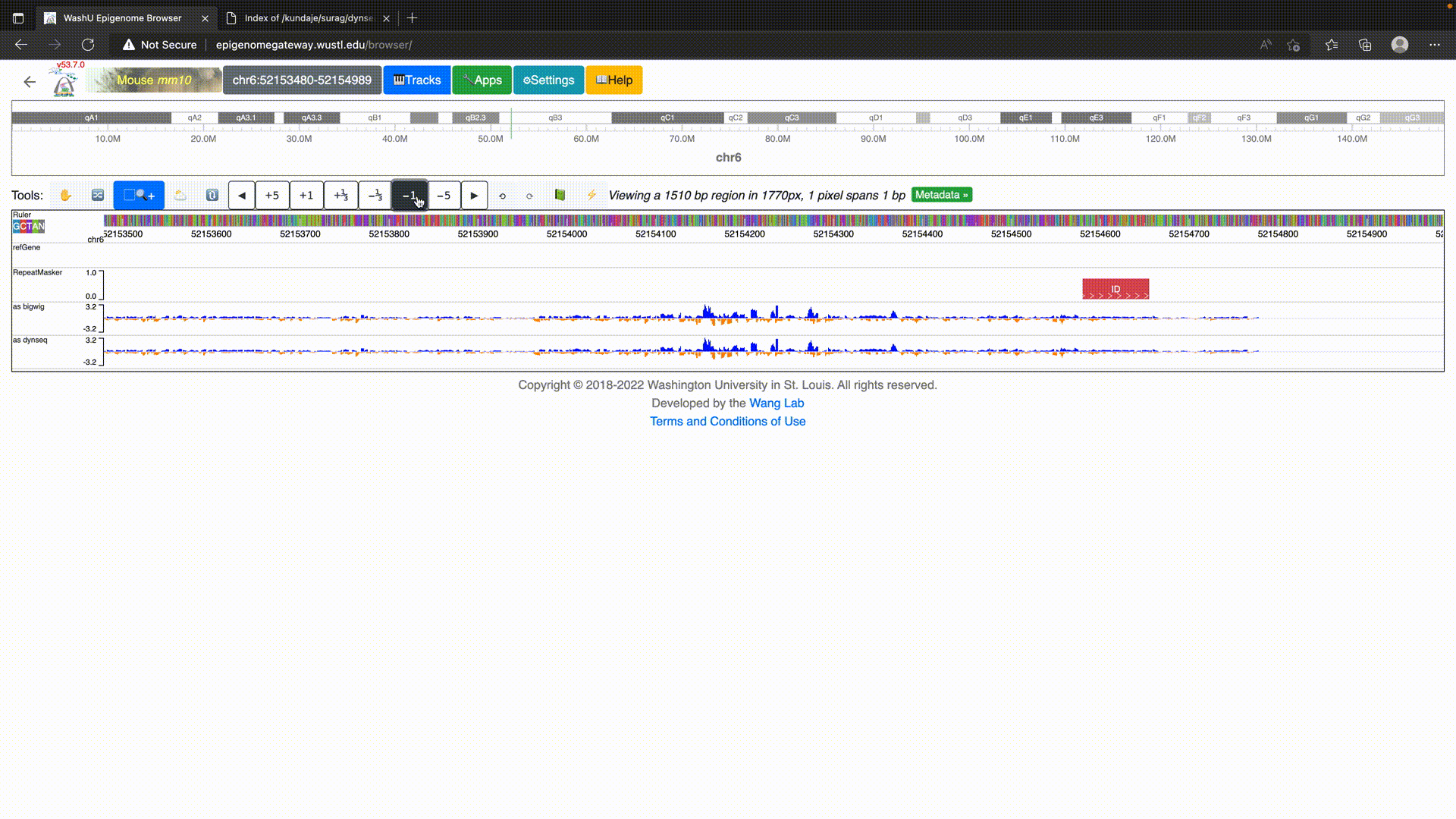
To use dynseq tracks on the WashU Epigenome Browser, simply load in the BigWig file using the “dynseq” track option as below.
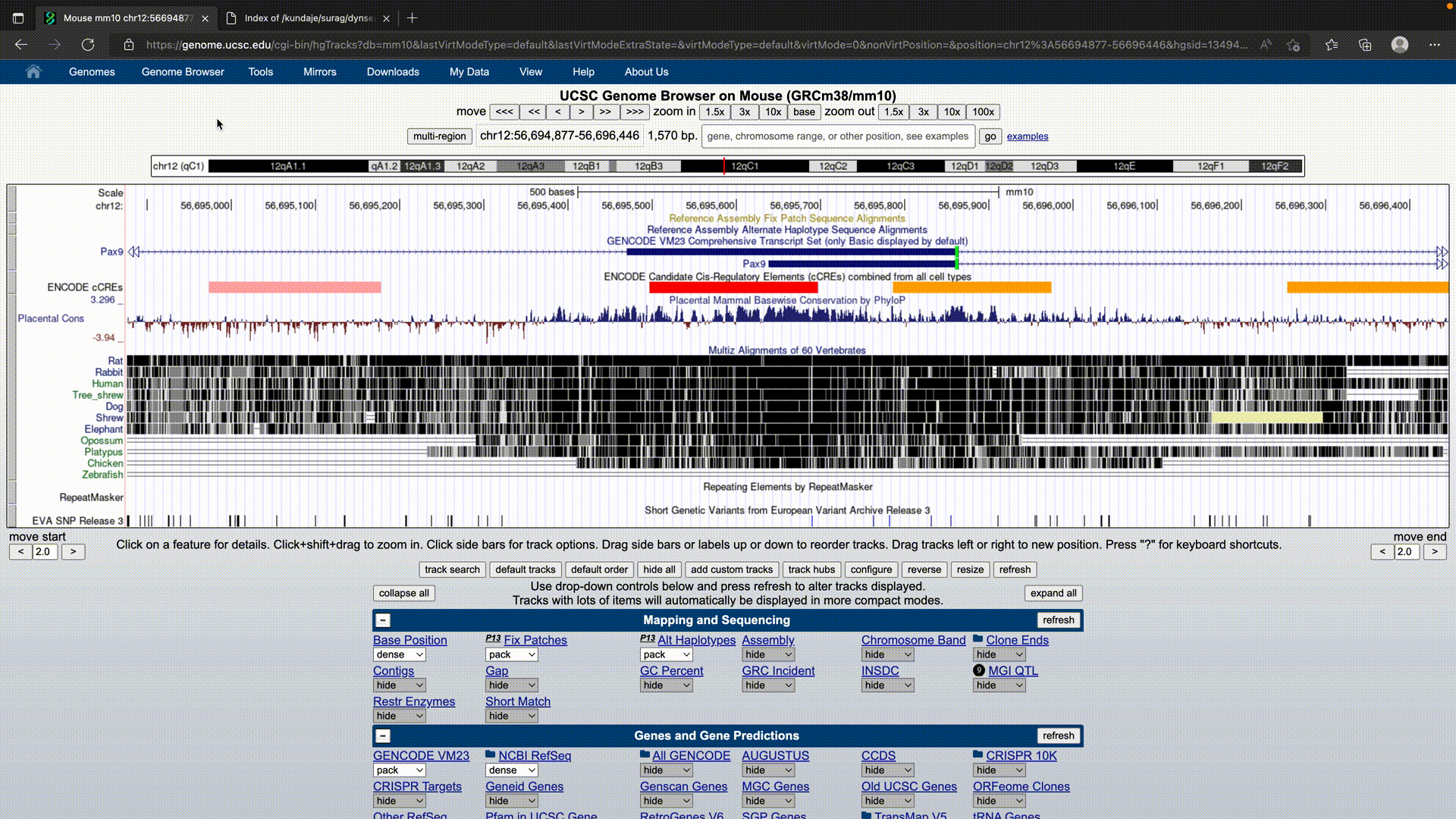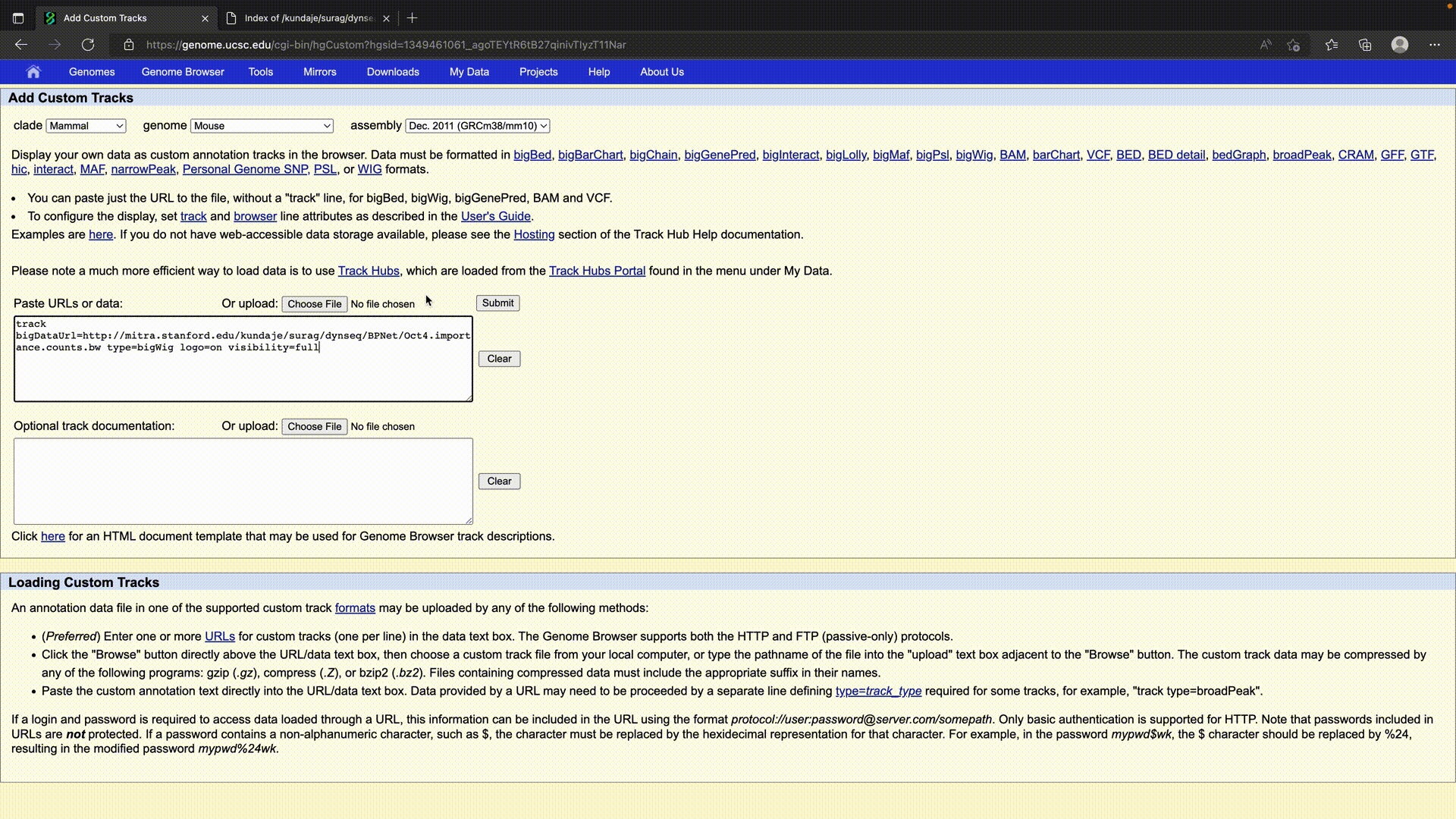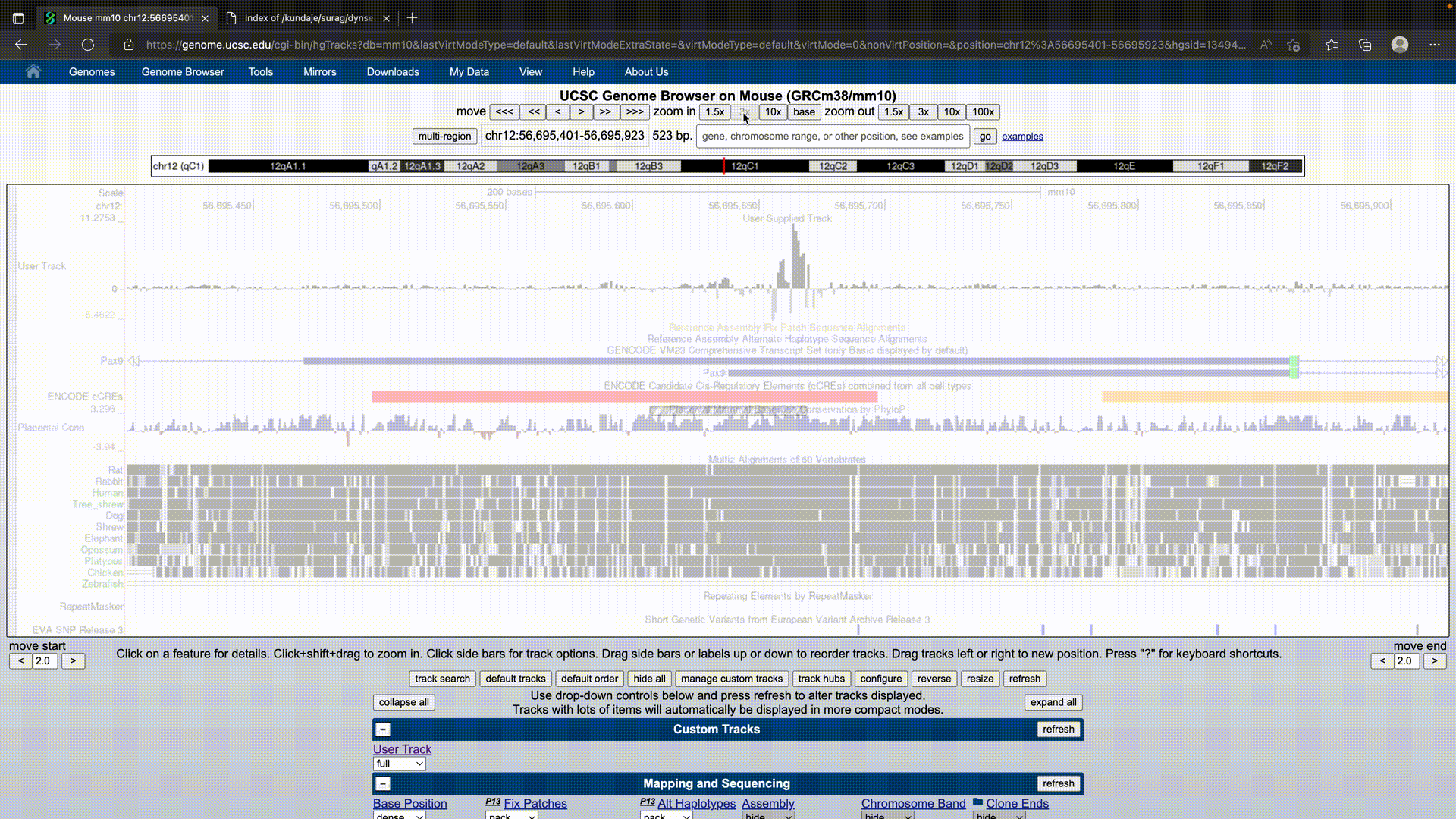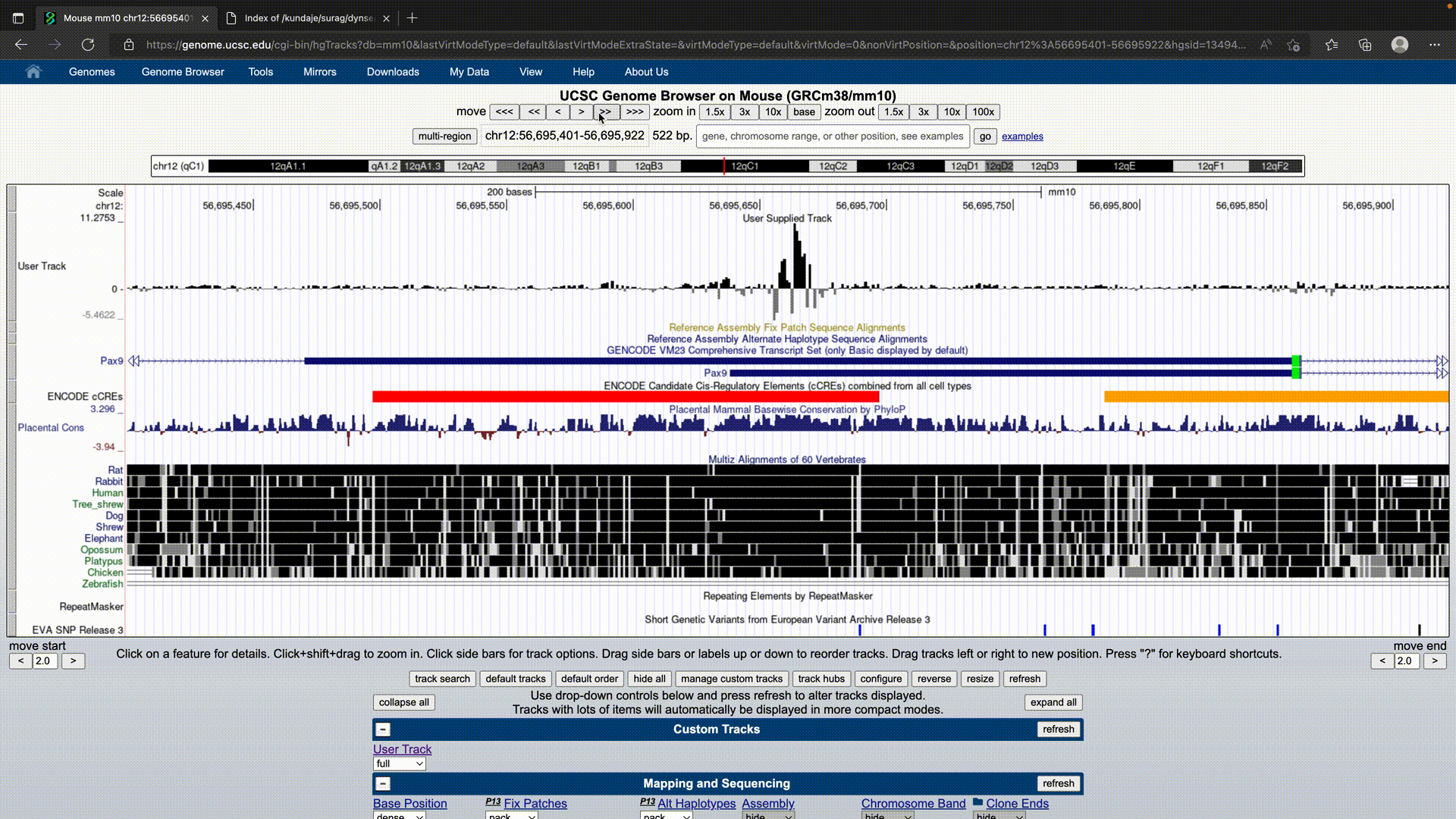
UCSC Genome Browser
The UCSC Browser treats the dynseq track as a regular BigWig that requires logo=on to be specified.
It can be specified as:
track bigDataUrl=https://link.to.dynseq.bw type=bigWig logo=on visibility=full
HiGlass/Resgen
The dynseq track has been implemented in HiGlass and supported by Resgen.
In HiGlass/Resgen, a dynseq track is associated with a BigWig and a Fasta file. This allows visualizing variants by providing an alternate Fasta file. To use dynseq in Resgen, first load the genome Fasta and chromosome size tsv with appropriate tags. Then add the dynseq track as shown below.
You can find the extended version which starts from loading the fasta and chromsizes file here.